📊 Results : 6GB rtx 3050 - 10 Epochs
Training Loss
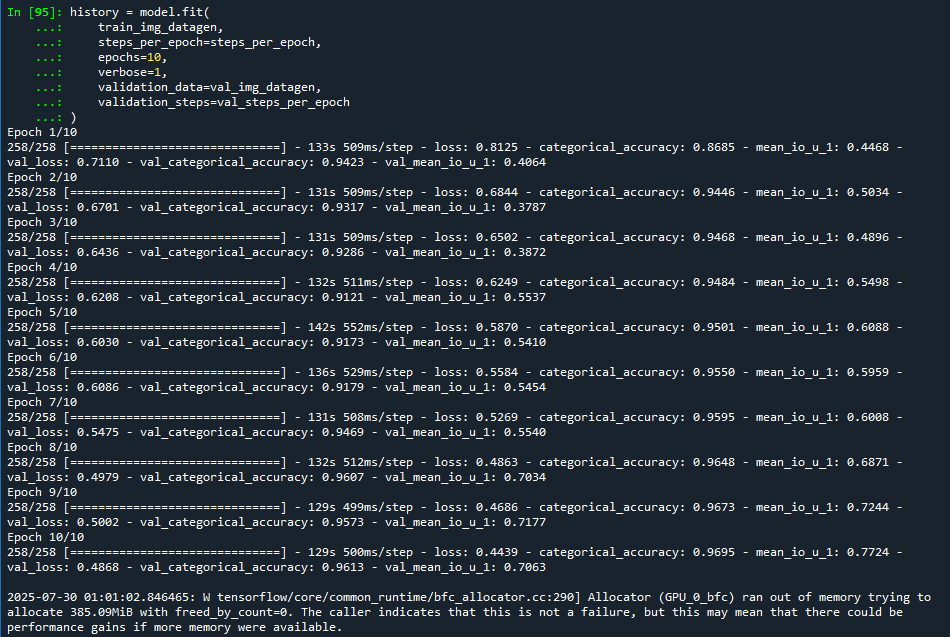
Mean IoU: 61.34 (10 Epochs)
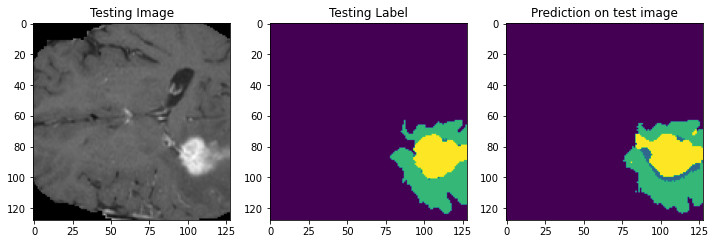
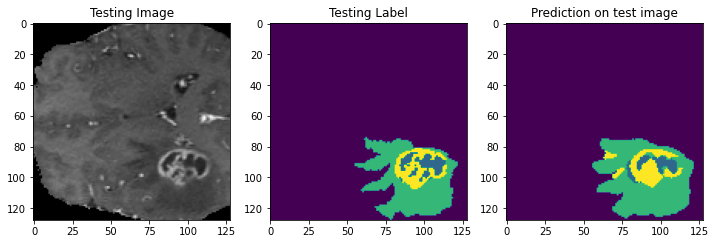
Prediction Samples
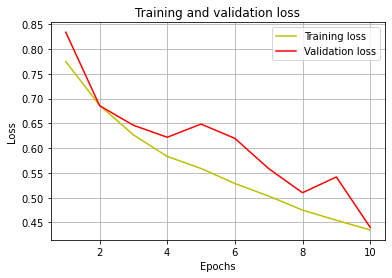
Training vs Validation Loss
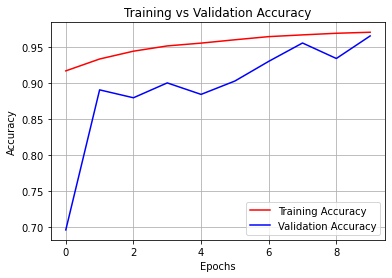
Training vs Validation Accuracy
The dataset used is the BraTS 2020 multimodal brain tumor segmentation dataset, available on Kaggle:
https://www.kaggle.com/datasets/awsaf49/brats20-dataset-training-validation
.nii.gz (NIfTI) format.0: Background1: Necrotic/Non-enhancing tumor core (NCR/NET)2: Peritumoral edema (ED)4: GD-enhancing tumor (ET)numpynibabelglobmatplotlibtensorflow and kerassklearn.preprocessing.MinMaxScalernibabelMinMaxScaler240×240×155 to 128×128×128 for uniform trainingsplitfoldersImageDataGenerator doesn't support .npy filesConv3D, MaxPooling3D, and UpSampling3Dmatplotlib (random 2D slices)



